Introduction to Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing, also known as antibiogram or antibiotic sensitivity testing, is a laboratory technique used to determine the sensitivity of a microorganism, such as bacteria, to particular antibiotics. This test helps doctors and clinicians determine the most appropriate antibiotic to treat a bacterial infection. When bacteria are exposed to an antibiotic, they can either be classified as Antimicrobial Susceptibility Testing, meaning the antibiotic can effectively kill the bacteria, or they are classified as resistant where the antibiotic has reduced or no ability to kill the bacteria. The results allow clinicians to optimize antibiotic therapy and minimize unnecessary or inappropriate use of antibiotics that would likely be ineffective.
Types of Antimicrobial Susceptibility Tests
There are a few different types of tests that are commonly used to determine the susceptibility of bacteria to various antibiotics:
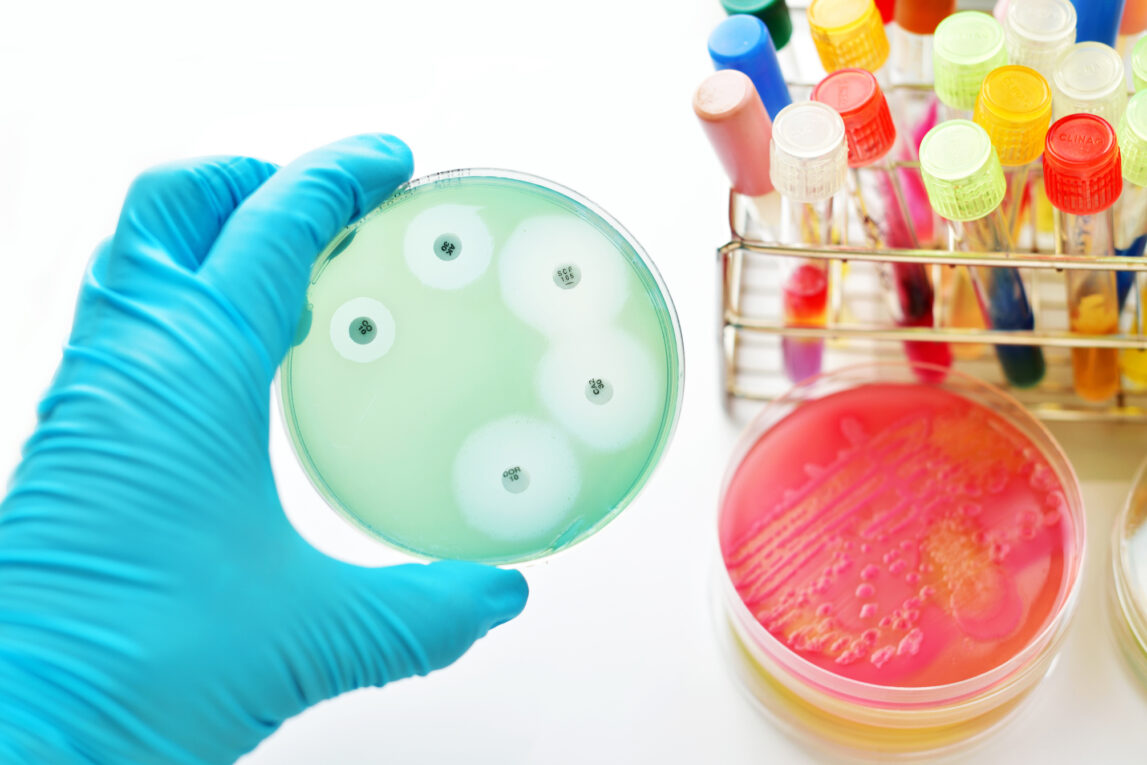
Disk diffusion tests – Also known as Kirby-Bauer testing, this involves applying filter paper disks impregnated with an antibiotic onto a culture plate inoculated with the bacterial isolate. As the antibiotic diffuses out into the agar, a gradient of decreasing concentration is established. If growth is inhibited around the disk, the bacteria are said to be susceptible to that antibiotic at normal dosing levels.
Automated broth microdilution – With this method, serial two-fold dilutions of antibiotics are prepared in microtitre plates. A standardized inoculum of the bacterial isolate is added to each well and indicators detect growth after incubation. The minimum inhibitory concentration, or lowest concentration that prevents growth, is determined for each antibiotic.
Agar dilution – As with broth microdilution, predefined dilutions of antibiotics are incorporated into agar plates. A standardized inoculum is spotted onto the surface and the plates are incubated. The lowest concentration that completely inhibits growth indicates susceptibility.
Etest – Polymeric strips containing a preformed antibiotic concentration gradient are applied to inoculated plates. Elliptical zones of inhibition are measured and interpreted based on clinical breakpoints.
These different testing approaches provide complementary information regarding antibiotic potency, but the disk diffusion and automated microdilution methods are most commonly used in clinical bacteriology laboratories. Standards ensure consistency in results across labs.
Reporting and Interpreting Antimicrobial Susceptibility Test Results
Once the tests are completed, results are reported quantitatively by determining the minimum inhibitory concentrations (MICs) required to inhibit growth. Qualitative categorizations of Antimicrobial Susceptibility Testing, intermediate, or resistant are also provided based on predefined clinical breakpoints established by organizations like the Clinical and Laboratory Standards Institute (CLSI).
Categories provide guidance on expected efficacy, with “susceptible” indicating the antibiotic can be reliably used for treatment at normal dosing. “Intermediate” means response may be uncertain and higher doses may be needed. “Resistant” implies the drug will likely not be effective as a single agent against that bug/drug combination. Labs also report epidemiological results like resistance frequencies within a facility.
Care must be taken when interpreting results, as many factors influence clinical outcome beyond just in vitro susceptibilities. Patient allergies, side effects, cost, pharmacokinetics, and site of infection must also be considered by clinicians. Repeat testing may also be needed if the patient fails to respond as expected or if the infection worsens. Together, test results along with clinical judgement allow optimization of antibiotic therapy whenever possible.
Common Bacteria Tested and Resistance Patterns
The most common bacterial pathogens routinely tested in clinical labs include Streptococcus pneumoniae, Staphylococcus aureus, Enterococcus spp., Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii. Periodic surveillance also tracks less frequently isolated species.
While a particular strain may be susceptible in the laboratory, antibiotic resistance continues rising globally due to overuse and misuse of these drugs. Multi-drug resistant organisms infecting hospitalized and immunocompromised patients remain a major public health concern. Carbapenem-resistant Enterobacteriaceae and multidrug-resistant Pseudomonas aeruginosa outbreaks highlight the urgent need for new antibiotics and antimicrobial stewardship to preserve our dwindling armamentarium.
Challenges and Limitations of Antimicrobial Susceptibility Testing
While susceptibility tests provide timely and clinically useful information, some limitations exist. Tests are performed on a pure cultured isolate, which may not fully reflect the susceptibility of all bacteria comprising a mixed infection. While accuracy is high, very minor errors in technique, reading zones of inhibition, or following interpretation guidelines could potentially lead to minor categorization disagreements between labs and even instruments.
Tests provide no information on achievable drug levels at the site of infection. Difficult-to-treat infections like endocarditis have different breakpoints than non-serious infections. Emergence of resistance during treatment cannot be predicted. New mechanisms of resistance may not be detected until breakpoints are re-established. Culture-negative specimens due to prior antibiotic exposure also limit the applicability of results.
Overall, antimicrobial susceptibility testing remains a cornerstone methodology for guiding antibiotic decisions. Continuous quality improvements, standardized methodologies, and integrated informatics solutions help maximize the clinical impact from these tests despite difficulties posed by mixed and culture-negative infections or changing resistance mechanisms over time.
*Note:
1. Source: Coherent Market Insights, Public sources, Desk research
2. We have leveraged AI tools to mine information and compile it.